I am very excited to say that my project for GSoC is complete! :)
I spent 5 days in Los Angeles at COMBINE 2014. I presented on August the 20th. Presentation link and video link are below.
Saturday, August 23, 2014
Tuesday, August 5, 2014
The Final Week is here!
Hello all,
I have quite a bit to report on the advancement of my project.
My current checklist was as so:
1. Complete synchronization of CellDesigner plugin model and JSBML.
2. maintain map of SBases.
2. Finish Render Extension additions
3. Work on adding cases to Reaction positioning algorithm(main reaction axes over 4 points were never handled, very diverse minority of cases)
During the last meeting, it was found that Rules, UnitDefinition, Events, Constraints, Parameters, etc are not synchronized in CellDesigner. Also, it was also found that IDs for many CellDesigner PluginSBases can be changed, and are not constant. Therefore, we could not depend on Ids to synchronize the CellDesigner and JSBML models.
As such, a new algorithm was proposed for Changing and Deleting SBases.
If a PluginSBase(other than Models or Compartments) is changed or deleted, we:
1. delete all associated JSBML objects in Model and Layout
2. delete them from the Map of SBases
2. re-add them directly from the CellDesigner model.
This has been done for all CellDesigner PluginSBases, and as such, there is a thorough synchronization.
The map is also maintained properly.
On the Layout front, I FINALLY got reaction axes over 4 points to be read properly by my algorithm. There are still small bugs that need to be fleshed out, but I am happy a few downloaded models actually render 95%+ properly.
The above model did not render properly until main reaction axes over four points were handled. This is an excellent way to assure conformance to models "in the wild".
Adding render information has always been difficult due to the complexity of the Render Extension itself, so I have not been getting very far on that. But I hope tomorrow will be a better day for that.
Finally, my practice presentation for the final presentation is going to be on the 13th in preparation for COMBINE on the 20th. I will need to start creating some demos to show off the progress I have made this summer on improving the JSBML/CellDesigner plugin interface.
I have quite a bit to report on the advancement of my project.
My current checklist was as so:
1. Complete synchronization of CellDesigner plugin model and JSBML.
2. maintain map of SBases.
2. Finish Render Extension additions
3. Work on adding cases to Reaction positioning algorithm(main reaction axes over 4 points were never handled, very diverse minority of cases)
During the last meeting, it was found that Rules, UnitDefinition, Events, Constraints, Parameters, etc are not synchronized in CellDesigner. Also, it was also found that IDs for many CellDesigner PluginSBases can be changed, and are not constant. Therefore, we could not depend on Ids to synchronize the CellDesigner and JSBML models.
As such, a new algorithm was proposed for Changing and Deleting SBases.
If a PluginSBase(other than Models or Compartments) is changed or deleted, we:
1. delete all associated JSBML objects in Model and Layout
2. delete them from the Map of SBases
2. re-add them directly from the CellDesigner model.
This has been done for all CellDesigner PluginSBases, and as such, there is a thorough synchronization.
The map is also maintained properly.
On the Layout front, I FINALLY got reaction axes over 4 points to be read properly by my algorithm. There are still small bugs that need to be fleshed out, but I am happy a few downloaded models actually render 95%+ properly.
Adding render information has always been difficult due to the complexity of the Render Extension itself, so I have not been getting very far on that. But I hope tomorrow will be a better day for that.
Finally, my practice presentation for the final presentation is going to be on the 13th in preparation for COMBINE on the 20th. I will need to start creating some demos to show off the progress I have made this summer on improving the JSBML/CellDesigner plugin interface.
Monday, July 28, 2014
Synchronizing CellDesigner and JSBML, Contact with Akira and Checklist
Just a quick update since it is late here, but over the past week, I have been continuing with my synchronization of the JSBML/CellDesigner(CD) model representations. I have added a Map<PluginSBase, Set<SBase>>. The key is the CD object and all JSBML objects associated with the CD object.
Since establishing contact with the main developer of CellDesigner (Dr. Akira Funahashi), I have sent him an email detailing some questions I had regarding CellDesigner.
My questions to him:
1. Is there is an accurate way to determine the reaction direction, be it North-South, East-West, etc? This answer will be very useful in determining the position of the Reaction text element, as it is calculated dynamically behind the scenes in CellDesigner.
2.According to the image below, when the CD user moves only a Reaction, nothing happens. One has to move a Species to send a Reaction SBaseChange event. This is unexpected and I would like some clarification on this if possible.
Current checklist for myself:
1. Add Render information
Since establishing contact with the main developer of CellDesigner (Dr. Akira Funahashi), I have sent him an email detailing some questions I had regarding CellDesigner.
My questions to him:
1. Is there is an accurate way to determine the reaction direction, be it North-South, East-West, etc? This answer will be very useful in determining the position of the Reaction text element, as it is calculated dynamically behind the scenes in CellDesigner.
2.According to the image below, when the CD user moves only a Reaction, nothing happens. One has to move a Species to send a Reaction SBaseChange event. This is unexpected and I would like some clarification on this if possible.
3. Some CellDesigner model files force CellDesigner to send SBaseChanged events every time one part of the model is changed. This is odd behavior, as if the whole model is changed when one small part is changed. It is even more odd considering as other models only send SBaseChanged events for PluginSBases that are edited by the user rather than an event for every PluginSBase in the model.
Current checklist for myself:
1. Add Render information
2. setCharge()/ FBC.
3. add reactiontext info, fix bugs
4. find species reference sbases in CellDesigner
Friday, July 18, 2014
Aftermath of 17/7 Meeting
I presented the slideshow below to Professor Funahashi (Akira), the main developer of CellDesigner, and my mentors at 15:00 UTC on 17/7/2014.
The presentation was a success because Akira was able to provide me with inside information and clarity to my work with CellDesigner. I asked questions at the end of the presentation which I will share answers with now:
Reaction text is calculated dynamically and hopefully, the algorithm will be provided to the team so we can also calculate reaction text in an identical way in JSBML.
PluginModel objects are not constant. As such, using PluginModels as a key in a Hashmap is impossible. Therefore, we need to remove our HashMap in AbstractCellDesignerPlugin and simply convert one model at a time.
I am pleased by the progress at this meeting and have high morale. I expect a lot of progress in the next few days.
Saturday, July 12, 2014
CellDesigner Reaction positioning and CellDesigner/JSBML synchronization
Over the past week, I have been working on two separate topics:
1. Complete algorithm that use the CellDesigner Plugin API to calculate positioning for ReactionGlyphs and SpeciesReferenceGlyphs.
2. Establish way to synchronize CellDesigner's plugin datastructure and JSBML (so that CellDesigner models will be immediately and accurately represented in JSBML)
The first point has been completed. ReactionGlyphs and SpeciesReferenceGlyph positioning has been accurately implemented. I still have loose ends to tie up, but I am proud that my algorithms do the "right thing" in a variety of situations.
On the second point, I have created a plugin that receives events from CellDesigner and displays them in a console. As you can see, when the position of a species or reaction changes, CellDesigner informs the console.
The next step is to update the currently selected model in JSBML automatically when it is edited by the user.
I did run into problems in the plugin implementation and I had a few small hang-ups in regards to getting consistent reaction coordinates, but things are progressing very well :)
-Ibrahim
1. Complete algorithm that use the CellDesigner Plugin API to calculate positioning for ReactionGlyphs and SpeciesReferenceGlyphs.
2. Establish way to synchronize CellDesigner's plugin datastructure and JSBML (so that CellDesigner models will be immediately and accurately represented in JSBML)
The first point has been completed. ReactionGlyphs and SpeciesReferenceGlyph positioning has been accurately implemented. I still have loose ends to tie up, but I am proud that my algorithms do the "right thing" in a variety of situations.
On the second point, I have created a plugin that receives events from CellDesigner and displays them in a console. As you can see, when the position of a species or reaction changes, CellDesigner informs the console.
The next step is to update the currently selected model in JSBML automatically when it is edited by the user.
I did run into problems in the plugin implementation and I had a few small hang-ups in regards to getting consistent reaction coordinates, but things are progressing very well :)
-Ibrahim
Wednesday, July 2, 2014
Calculating ReactionGlyph position in CellDesigner and new Plugin
CellDesigner does not make its Reaction position data available for developers to access. Therefore, we need to figure out how to determine Reaction size and position using our own algorithms.
We have two choices:
1. Create a BoundingBox with x,y, height and width.
2. Create a Curve for both the ReactionGlyph and the SpeciesReferenceGlyphs.
-ReactionGlyph curve segment will be in the middle of the Reaction.
-SpeciesReferenceGlyph curve segments will be coming off the center curve segment and pointing to SpeciesGlyphs, be it product, reactant, or modifier.
The image above shows the difference between a ReactionGlyph and a SpeciesReferenceGlyph. The ReactionGlyph is the entire Reaction shape, while there are three SpeciesReferenceGlyphs which are separated by the small black circle, each leading to a SpeciesGlyph.
Reaction position would be trivial if all reactions looked like the one above. However, reactions that make up a CellDesigner model can come in any and all orientations and positions, like the image below.
As one can see, reactants, products, and modifiers are in all orientations respective to one another, which makes it very difficult to produce accurate position measurements. I have been working on this problem for a few days and still working on it.
While doing this, I have also started on writing a plugin that prints out CellDesigner information(PropertyChangeEvents). This also does not work as of now. However, I have my weekly group meeting tomorrow at 15:00 UTC which will hopefully put me on the right track regarding these issues.
We have two choices:
1. Create a BoundingBox with x,y, height and width.
2. Create a Curve for both the ReactionGlyph and the SpeciesReferenceGlyphs.
-ReactionGlyph curve segment will be in the middle of the Reaction.
-SpeciesReferenceGlyph curve segments will be coming off the center curve segment and pointing to SpeciesGlyphs, be it product, reactant, or modifier.
The image above shows the difference between a ReactionGlyph and a SpeciesReferenceGlyph. The ReactionGlyph is the entire Reaction shape, while there are three SpeciesReferenceGlyphs which are separated by the small black circle, each leading to a SpeciesGlyph.
Reaction position would be trivial if all reactions looked like the one above. However, reactions that make up a CellDesigner model can come in any and all orientations and positions, like the image below.
While doing this, I have also started on writing a plugin that prints out CellDesigner information(PropertyChangeEvents). This also does not work as of now. However, I have my weekly group meeting tomorrow at 15:00 UTC which will hopefully put me on the right track regarding these issues.
Thursday, June 26, 2014
Midterm Presentation
Here are my slides that I presented today for the midterm evaluation meeting:
Dr. Hucka linked me to some Layout Extension links that I should look into further. They are here:
"* Frank wrote an online SBML layout viewer: http://sysbioapps.dyndns.org/
* COPASI should support reading layout: http://frank-fbergmann.
* There are more tools that mention SBML layout support, at http://sbml.org/SBML_Software_
Things are going well with my project. These are my future plans:
Dr. Hucka linked me to some Layout Extension links that I should look into further. They are here:
"* Frank wrote an online SBML layout viewer: http://sysbioapps.dyndns.org/
* COPASI should support reading layout: http://frank-fbergmann.
* There are more tools that mention SBML layout support, at http://sbml.org/SBML_Software_
Things are going well with my project. These are my future plans:
Saturday, June 21, 2014
Preparing for the Second Group Meeting and Project Update
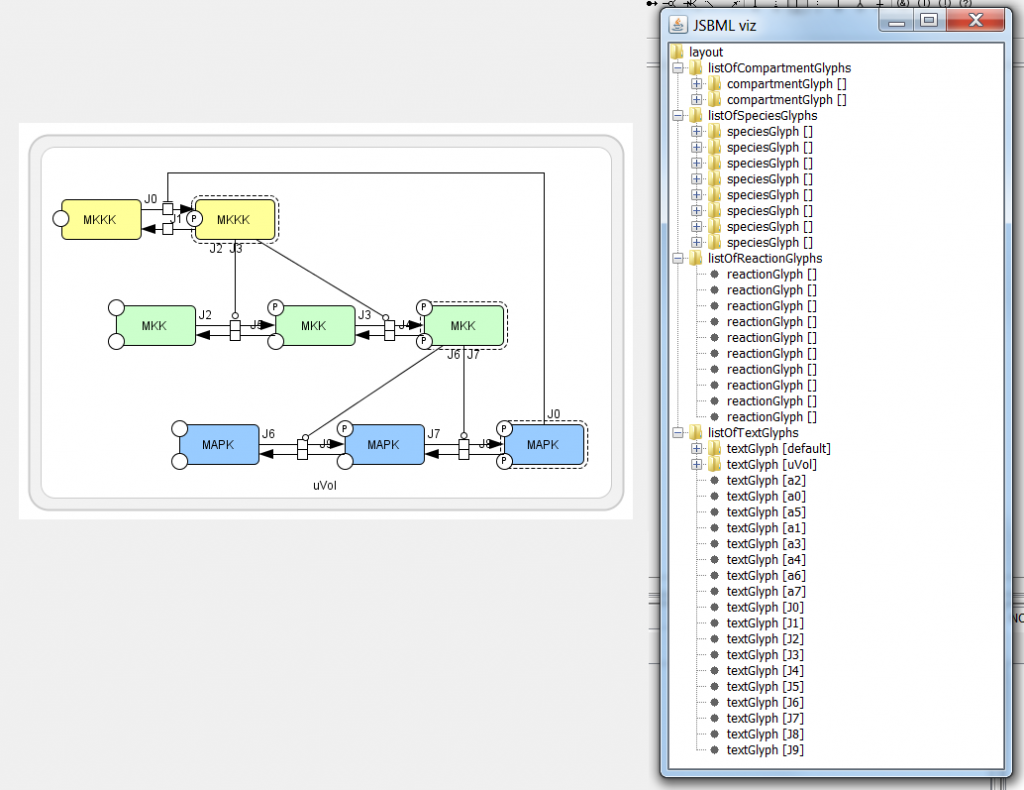
After 4 hours of struggle and toil debugging my code and figuring out why CellDesigner's plugin interface was crashing: the photo below is the result. Without them, I would not have been able to work through the myriad of errors and obstacles that were appearing. Some of these errors had to do with the fact that one null pointer or any oversight caused no observable error message, but simply a dramatic failure, which is analogous to a missing nail collapsing the Empire State Building. Luckily, my mentors and I cobbled together a class that gives lots of debugging information when a plugin crashes.
As one can see from the image, TextGlyphs, ReactionGlyphs, CompartmentGlyphs, and SpeciesGlyphs have now been added to the JSBML Layout Package. This is a great first step to completing my project's aims.
The Second Group Meeting and my second presentation is on Wednesday the 25th. My first presentation went well, and I have plenty to talk about for the second presentation. I will post my presentation here soon.
As one can see from the image, TextGlyphs, ReactionGlyphs, CompartmentGlyphs, and SpeciesGlyphs have now been added to the JSBML Layout Package. This is a great first step to completing my project's aims.
The Second Group Meeting and my second presentation is on Wednesday the 25th. My first presentation went well, and I have plenty to talk about for the second presentation. I will post my presentation here soon.
Sunday, June 15, 2014
At the Cusp of Great Progress
Hello everyone,
I just committed a revision to the JSBML project, most likely the biggest since I began committing JUnit tests in early May. I:
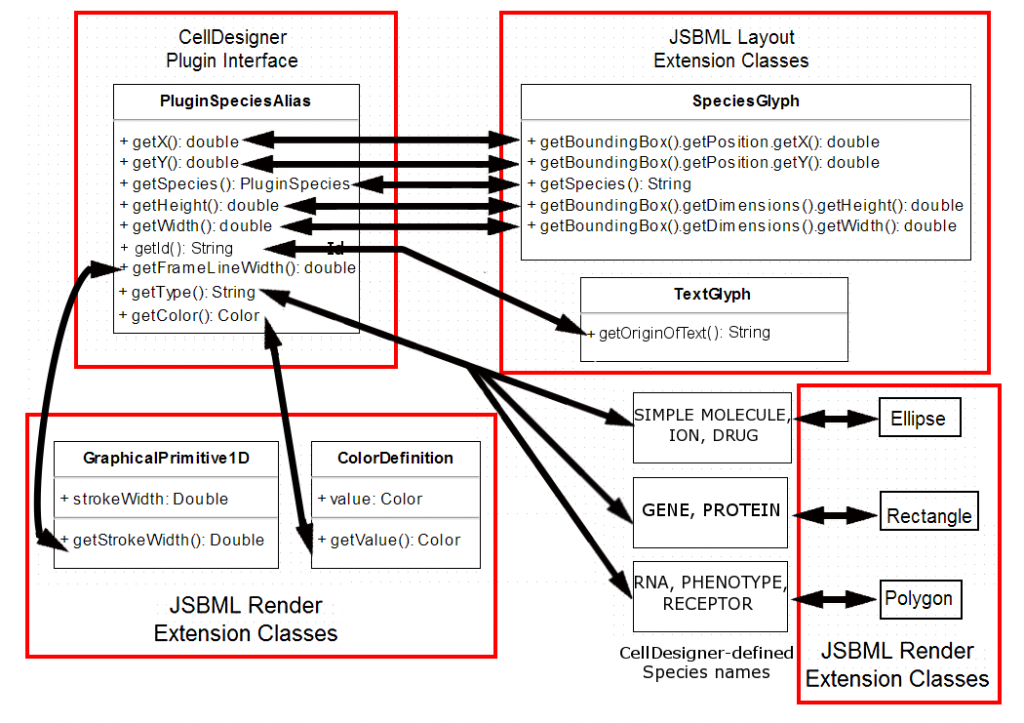
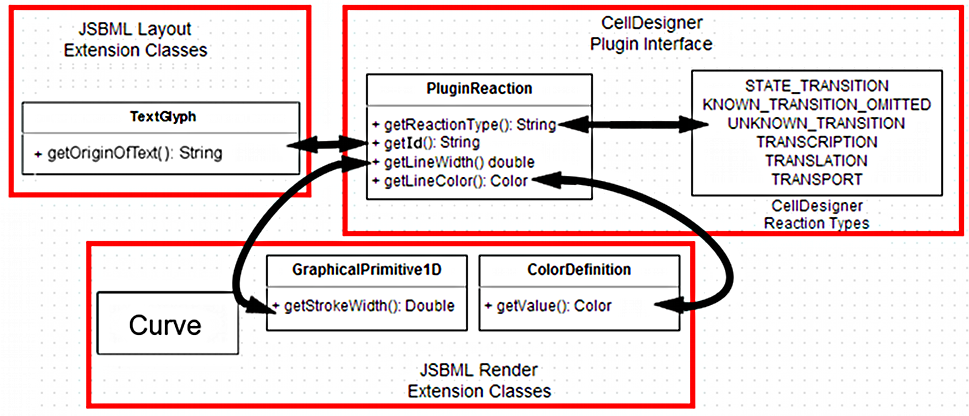
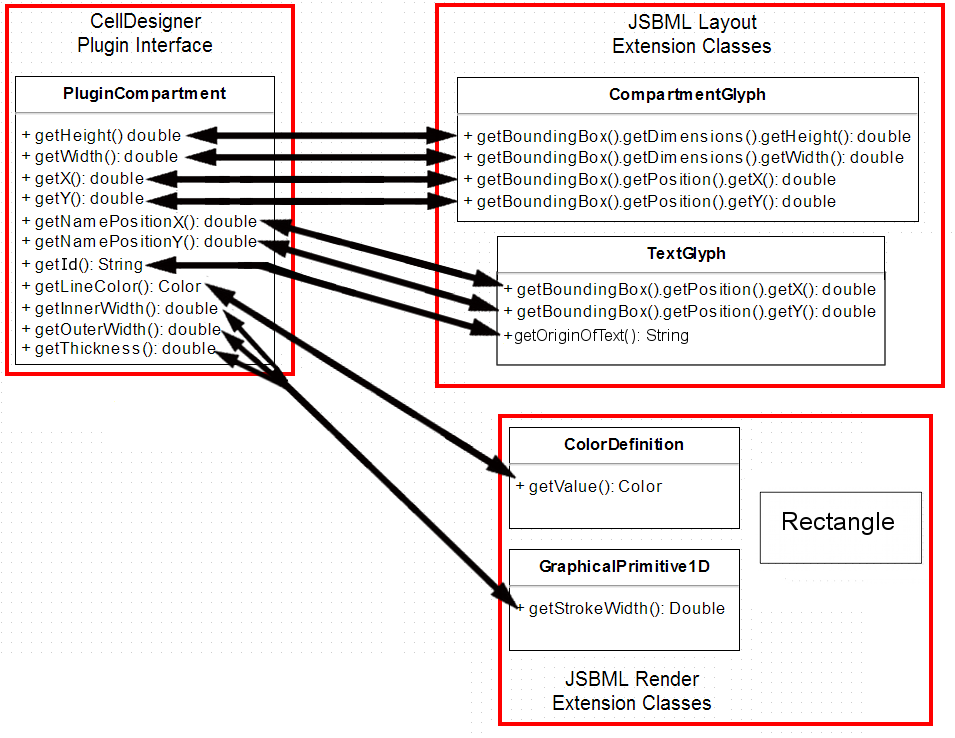
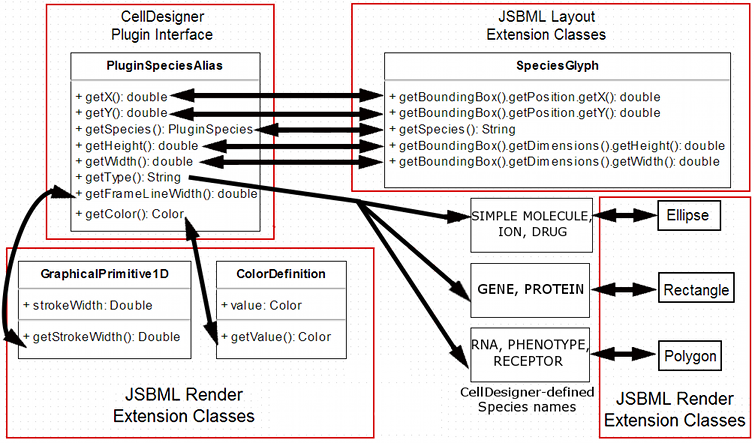
I have also been developing schematics for my main project, the mapping of CellDesigner data structures to JSBML data structures. They are shown below in their (probably) final form.
1. SpeciesGlyph
2. ReactionGlyph
I just committed a revision to the JSBML project, most likely the biggest since I began committing JUnit tests in early May. I:
- Added two more JUnit Tests to the Render Package
- Added UserMessages.xml in the examples/resource folder to the CellDesigner build.xml file
- Moved SwingWorker to its' own file
- AbstractCellDesignerPlugin now implements PropertyChangeListener
I did run into problems with the Ant Script in regards to adding UserMessages.xml to the CellDesigner build file. I will be asking my mentors about how to fix this issue.
I have also been developing schematics for my main project, the mapping of CellDesigner data structures to JSBML data structures. They are shown below in their (probably) final form.
1. SpeciesGlyph
2. ReactionGlyph
3. CompartmentGlyph
Current To-Do List
- Suggest additions to user guide setting up JSBML development in Eclipse.
- Fix UserMessages.xml problem.
- Begin to convert the CellDesigner plugin data structure to JSBML data structure via a new class called convertModelLayoutPackage (or something analogous)
Monday, June 2, 2014
Render Package JUnit Tests and Schematics
May 28th was our Group Meeting, and Victor and Leandro did excellent work! :)
Over the past week, I have been working on extending the coverage of the JUnit tests in the Render Extension Package. There are now 10 completed unit tests out of the 34 Render Classes.
I will be making other schematics for classes in the Layout Package ReactionGlyph, TextGlyph, CompartmentGlyph, etc.
I have not been running into any problems that I have not been able to solve this week.
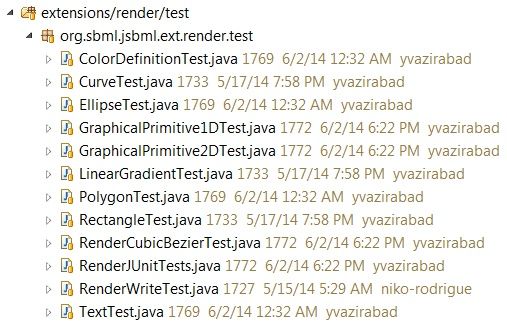 |
| The Render Package JUnit tests as of June 2nd, 2014 |
I am planning on making schematics of each mapping of the CellDesigner plugin interface. The image below is the first schematic of a series. It was first seen during the May 28th Presentation.
I will be making other schematics for classes in the Layout Package ReactionGlyph, TextGlyph, CompartmentGlyph, etc.
I have not been running into any problems that I have not been able to solve this week.
I will continue my JUnit testing, and I will begin to:
Develop CellDesigner plugin.
Develop CellDesigner plugin.
1.Obtains CellDesigners's plugin
structure and export pure Layout/Render
SBML.
2.Visualization of SBML
network changes via JTree.
—Synchronize changes
between plugin and CellDesigner.
Labels:
API,
CellDesigner,
JSBML,
JUnit
Location:
Milwaukee, WI, USA
Tuesday, May 27, 2014
Presentation at 17:00 UTC on 28/5/2014. Be there or be Square!
Hello all,
Sorry for not blogging lately. I have been busy with sorting out my project goals and creating my presentation for the group meeting at 17:00 UTC on 28/5/2014.
So, without further ado, here is my presentation!
Sorry for not blogging lately. I have been busy with sorting out my project goals and creating my presentation for the group meeting at 17:00 UTC on 28/5/2014.
So, without further ado, here is my presentation!
Saturday, May 17, 2014
Render Package Testing
Over the past week, I have been doing a few things. The main task was to develop JUnit test files for specific classes in the Render Extension package.
The Render Specification was perused and then these classes were tested:
1. ColorDefinition
2. Curve
3. Ellipse
4. LinearGradient
5. Polygon
6. Rectangle
7. Text
I encountered one problem in developing these tests. For some reason, I was getting NullPointerExceptions for the getcx, setcx, or issetcx methods of the Ellipse class, even though the other cy and cz methods were fine. It was inexplicable. So I initialized the cx variable in the Ellipse class to 0d and the methods then worked.
I need to get ready for the next joint meeting on May 28th at 5 PM UTC(12 PM CST). I am already making preliminary preparations.
I still need to read the CellDesigner Extension Tag Specification Document. I will most likely read this on Sunday the 18th.
The Render Specification was perused and then these classes were tested:
1. ColorDefinition
2. Curve
3. Ellipse
4. LinearGradient
5. Polygon
6. Rectangle
7. Text
I encountered one problem in developing these tests. For some reason, I was getting NullPointerExceptions for the getcx, setcx, or issetcx methods of the Ellipse class, even though the other cy and cz methods were fine. It was inexplicable. So I initialized the cx variable in the Ellipse class to 0d and the methods then worked.
I need to get ready for the next joint meeting on May 28th at 5 PM UTC(12 PM CST). I am already making preliminary preparations.
I still need to read the CellDesigner Extension Tag Specification Document. I will most likely read this on Sunday the 18th.
Sunday, May 11, 2014
Systems Biology Graphical Notation
I have been reading the pertinent literature in regards to two portions of the Systems Biology Graphical Notation specifications:
1. Process Description Language
2. Activity Flow Language
Let us discuss each in turn.
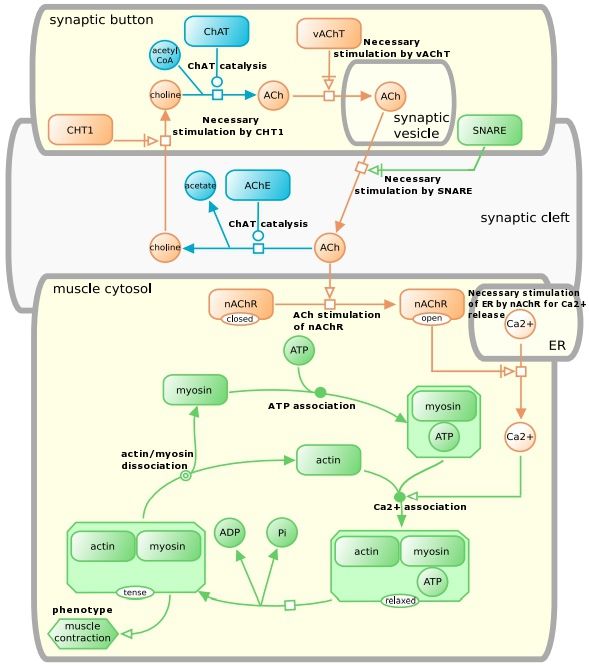
According to the SBGN website, the "Process Description language shows the temporal courses of biochemical interactions in a network. It can be used to show all the molecular interactions taking place in a network of biochemical entities, with the same entity appearing multiple times in the same diagram".
Let us examine an example process description map(from page 57 of the specifications PDF) using the above reference card.
Onto the Activity Flow language specification:
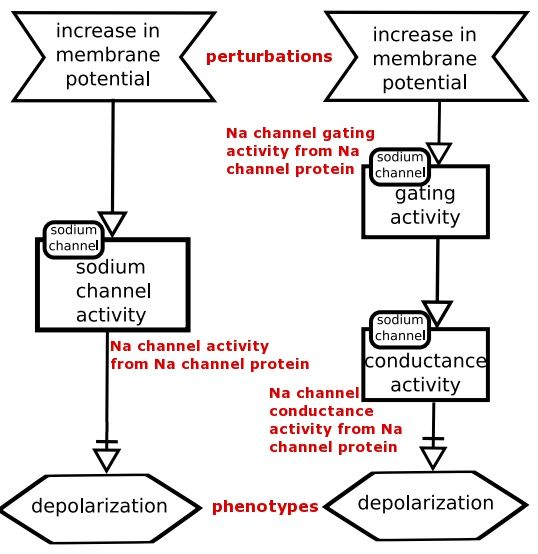
The Activity Flow Language "depicts the flow of information between biochemical entities in a network. It omits information about the state transitions of entities and is particularly convenient for representing the effects of perturbations, whether genetic or environmental in nature".
An Activity Flow Description map is annotated below.
1. Process Description Language
2. Activity Flow Language
Let us discuss each in turn.
According to the SBGN website, the "Process Description language shows the temporal courses of biochemical interactions in a network. It can be used to show all the molecular interactions taking place in a network of biochemical entities, with the same entity appearing multiple times in the same diagram".
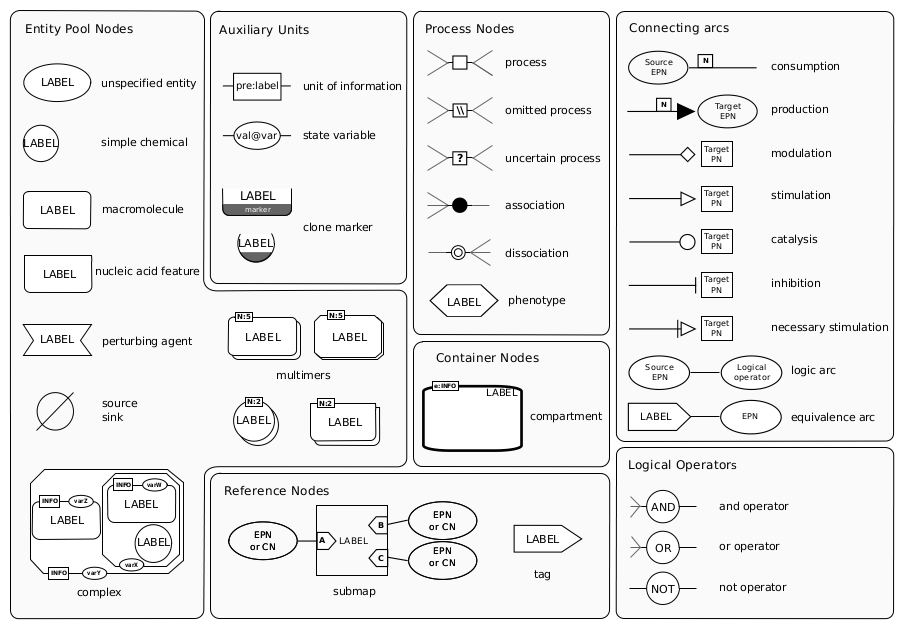 |
| Reference Card for SBGN symbols |
The Activity Flow Language "depicts the flow of information between biochemical entities in a network. It omits information about the state transitions of entities and is particularly convenient for representing the effects of perturbations, whether genetic or environmental in nature".
An Activity Flow Description map is annotated below.
Location:
Milwaukee, WI, USA
Friday, May 9, 2014
On to Week 3!
GSoC has been going on for over 2 weeks, and the JSBML team and I have been getting to know one another through dozens of emails and a handful of Google Hangouts. Since this was my first time working with a highly established project such as JSBML, there was a definite learning curve associated with setting up the project in Eclipse, setting up SVN/Subclipse, and checking out the project using https:// rather than http://. Dr. Dräger and Alex were incredibly patient with me and my workspace is finally up to snuff.
Since my proposal focuses on the CellDesigner module and the Layout/Render extension packages, my first assignment was to test and analyze an example CellDesigner plugin to understand the interplay between JSBML and the CellDesigner program. That being completed, I then was asked to develop two JUnit test classes for the SpeciesReferenceGlyph and TextGlyph classes which tested all get(), set() and isSet() methods. Then, a frankly elegant method for running all JUnit tests was suggested by Dr. Dräger and Dr. Rodriguez which is seen below.
When this class is inserted into the build script, it runs all JUnit tests which are inserted into the @SuiteClasses method.
My current assignment is to read both the Activity Flow Language specification and the Process Description Language specification on the SBGN website.
Since my proposal focuses on the CellDesigner module and the Layout/Render extension packages, my first assignment was to test and analyze an example CellDesigner plugin to understand the interplay between JSBML and the CellDesigner program. That being completed, I then was asked to develop two JUnit test classes for the SpeciesReferenceGlyph and TextGlyph classes which tested all get(), set() and isSet() methods. Then, a frankly elegant method for running all JUnit tests was suggested by Dr. Dräger and Dr. Rodriguez which is seen below.
package org.sbml.jsbml.ext.layout;
import org.junit.runner.RunWith;
import org.junit.runners.Suite;
import org.junit.runners.Suite.SuiteClasses;
@RunWith(value=Suite.class)
@SuiteClasses(value={TextGlyphTest.class, SpeciesReferenceGlyphTest.class})
public class LayoutJUnitTests {
}
When this class is inserted into the build script, it runs all JUnit tests which are inserted into the @SuiteClasses method.
My current assignment is to read both the Activity Flow Language specification and the Process Description Language specification on the SBGN website.
Thursday, May 8, 2014
Introduction
Hello all!
My name is Ibrahim Vazirabad. I hold a bachelor's degree in BioMolecular Engineering from the Milwaukee School of Engineering and intend on starting my Master's degree in Bioinformatics at Marquette University starting this fall. This summer, I am being mentored by Dr. Andreas Dräger and Alex Thomas, both part of the JSBML community. My project this summer focuses on the improvement of the plug-in interface of CellDesigner and mapping the interface fully to JSBML data structures. This will assist in the porting of CellDesigner internals to JSBML from libSBML.
I am very enthusiastic about the opportunity to work with Dr. Dräger, Alex, and the rest of the JSBML team. I am sure that it will be a great summer.
My name is Ibrahim Vazirabad. I hold a bachelor's degree in BioMolecular Engineering from the Milwaukee School of Engineering and intend on starting my Master's degree in Bioinformatics at Marquette University starting this fall. This summer, I am being mentored by Dr. Andreas Dräger and Alex Thomas, both part of the JSBML community. My project this summer focuses on the improvement of the plug-in interface of CellDesigner and mapping the interface fully to JSBML data structures. This will assist in the porting of CellDesigner internals to JSBML from libSBML.
I am very enthusiastic about the opportunity to work with Dr. Dräger, Alex, and the rest of the JSBML team. I am sure that it will be a great summer.
Labels:
2014,
CellDesigner,
Google,
GSoC,
JSBML
Location:
Milwaukee, WI, USA
Subscribe to:
Posts (Atom)