Hello all,
Sorry for not blogging lately. I have been busy with sorting out my project goals and creating my presentation for the group meeting at 17:00 UTC on 28/5/2014.
So, without further ado, here is my presentation!
Tuesday, May 27, 2014
Saturday, May 17, 2014
Render Package Testing
Over the past week, I have been doing a few things. The main task was to develop JUnit test files for specific classes in the Render Extension package.
The Render Specification was perused and then these classes were tested:
1. ColorDefinition
2. Curve
3. Ellipse
4. LinearGradient
5. Polygon
6. Rectangle
7. Text
I encountered one problem in developing these tests. For some reason, I was getting NullPointerExceptions for the getcx, setcx, or issetcx methods of the Ellipse class, even though the other cy and cz methods were fine. It was inexplicable. So I initialized the cx variable in the Ellipse class to 0d and the methods then worked.
I need to get ready for the next joint meeting on May 28th at 5 PM UTC(12 PM CST). I am already making preliminary preparations.
I still need to read the CellDesigner Extension Tag Specification Document. I will most likely read this on Sunday the 18th.
The Render Specification was perused and then these classes were tested:
1. ColorDefinition
2. Curve
3. Ellipse
4. LinearGradient
5. Polygon
6. Rectangle
7. Text
I encountered one problem in developing these tests. For some reason, I was getting NullPointerExceptions for the getcx, setcx, or issetcx methods of the Ellipse class, even though the other cy and cz methods were fine. It was inexplicable. So I initialized the cx variable in the Ellipse class to 0d and the methods then worked.
I need to get ready for the next joint meeting on May 28th at 5 PM UTC(12 PM CST). I am already making preliminary preparations.
I still need to read the CellDesigner Extension Tag Specification Document. I will most likely read this on Sunday the 18th.
Sunday, May 11, 2014
Systems Biology Graphical Notation
I have been reading the pertinent literature in regards to two portions of the Systems Biology Graphical Notation specifications:
1. Process Description Language
2. Activity Flow Language
Let us discuss each in turn.
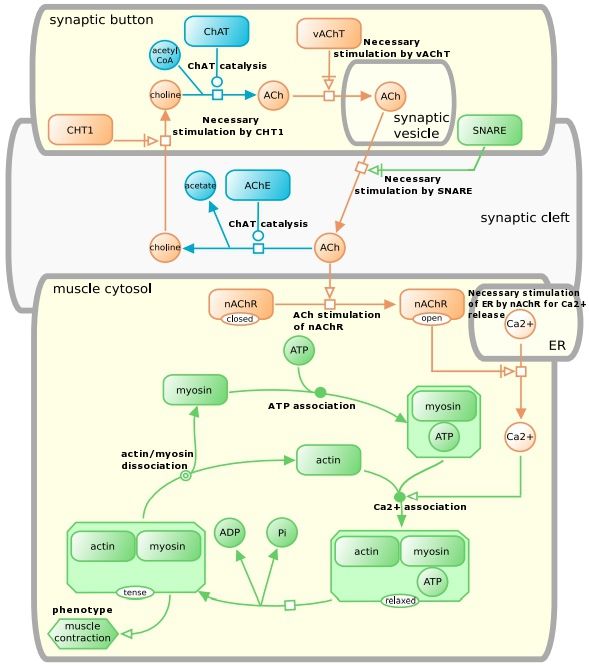
According to the SBGN website, the "Process Description language shows the temporal courses of biochemical interactions in a network. It can be used to show all the molecular interactions taking place in a network of biochemical entities, with the same entity appearing multiple times in the same diagram".
Let us examine an example process description map(from page 57 of the specifications PDF) using the above reference card.
Onto the Activity Flow language specification:
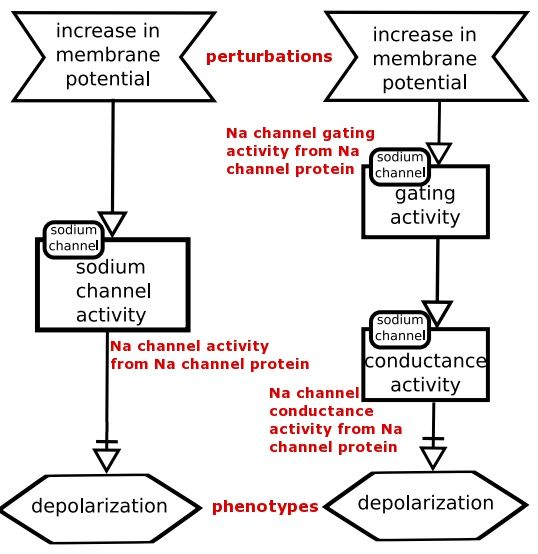
The Activity Flow Language "depicts the flow of information between biochemical entities in a network. It omits information about the state transitions of entities and is particularly convenient for representing the effects of perturbations, whether genetic or environmental in nature".
An Activity Flow Description map is annotated below.
1. Process Description Language
2. Activity Flow Language
Let us discuss each in turn.
According to the SBGN website, the "Process Description language shows the temporal courses of biochemical interactions in a network. It can be used to show all the molecular interactions taking place in a network of biochemical entities, with the same entity appearing multiple times in the same diagram".
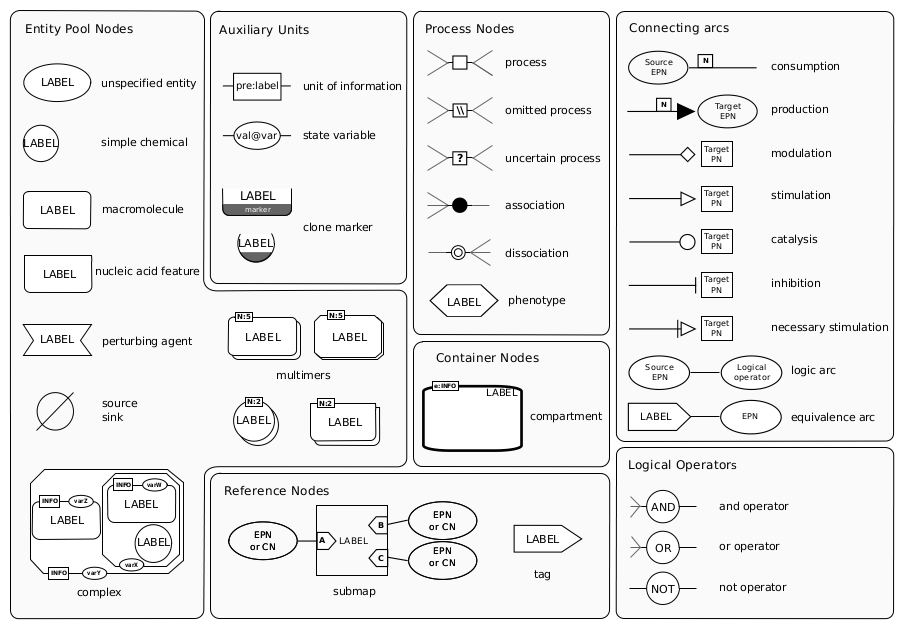 |
| Reference Card for SBGN symbols |
The Activity Flow Language "depicts the flow of information between biochemical entities in a network. It omits information about the state transitions of entities and is particularly convenient for representing the effects of perturbations, whether genetic or environmental in nature".
An Activity Flow Description map is annotated below.
Location:
Milwaukee, WI, USA
Friday, May 9, 2014
On to Week 3!
GSoC has been going on for over 2 weeks, and the JSBML team and I have been getting to know one another through dozens of emails and a handful of Google Hangouts. Since this was my first time working with a highly established project such as JSBML, there was a definite learning curve associated with setting up the project in Eclipse, setting up SVN/Subclipse, and checking out the project using https:// rather than http://. Dr. Dräger and Alex were incredibly patient with me and my workspace is finally up to snuff.
Since my proposal focuses on the CellDesigner module and the Layout/Render extension packages, my first assignment was to test and analyze an example CellDesigner plugin to understand the interplay between JSBML and the CellDesigner program. That being completed, I then was asked to develop two JUnit test classes for the SpeciesReferenceGlyph and TextGlyph classes which tested all get(), set() and isSet() methods. Then, a frankly elegant method for running all JUnit tests was suggested by Dr. Dräger and Dr. Rodriguez which is seen below.
When this class is inserted into the build script, it runs all JUnit tests which are inserted into the @SuiteClasses method.
My current assignment is to read both the Activity Flow Language specification and the Process Description Language specification on the SBGN website.
Since my proposal focuses on the CellDesigner module and the Layout/Render extension packages, my first assignment was to test and analyze an example CellDesigner plugin to understand the interplay between JSBML and the CellDesigner program. That being completed, I then was asked to develop two JUnit test classes for the SpeciesReferenceGlyph and TextGlyph classes which tested all get(), set() and isSet() methods. Then, a frankly elegant method for running all JUnit tests was suggested by Dr. Dräger and Dr. Rodriguez which is seen below.
package org.sbml.jsbml.ext.layout;
import org.junit.runner.RunWith;
import org.junit.runners.Suite;
import org.junit.runners.Suite.SuiteClasses;
@RunWith(value=Suite.class)
@SuiteClasses(value={TextGlyphTest.class, SpeciesReferenceGlyphTest.class})
public class LayoutJUnitTests {
}
When this class is inserted into the build script, it runs all JUnit tests which are inserted into the @SuiteClasses method.
My current assignment is to read both the Activity Flow Language specification and the Process Description Language specification on the SBGN website.
Thursday, May 8, 2014
Introduction
Hello all!
My name is Ibrahim Vazirabad. I hold a bachelor's degree in BioMolecular Engineering from the Milwaukee School of Engineering and intend on starting my Master's degree in Bioinformatics at Marquette University starting this fall. This summer, I am being mentored by Dr. Andreas Dräger and Alex Thomas, both part of the JSBML community. My project this summer focuses on the improvement of the plug-in interface of CellDesigner and mapping the interface fully to JSBML data structures. This will assist in the porting of CellDesigner internals to JSBML from libSBML.
I am very enthusiastic about the opportunity to work with Dr. Dräger, Alex, and the rest of the JSBML team. I am sure that it will be a great summer.
My name is Ibrahim Vazirabad. I hold a bachelor's degree in BioMolecular Engineering from the Milwaukee School of Engineering and intend on starting my Master's degree in Bioinformatics at Marquette University starting this fall. This summer, I am being mentored by Dr. Andreas Dräger and Alex Thomas, both part of the JSBML community. My project this summer focuses on the improvement of the plug-in interface of CellDesigner and mapping the interface fully to JSBML data structures. This will assist in the porting of CellDesigner internals to JSBML from libSBML.
I am very enthusiastic about the opportunity to work with Dr. Dräger, Alex, and the rest of the JSBML team. I am sure that it will be a great summer.
Labels:
2014,
CellDesigner,
Google,
GSoC,
JSBML
Location:
Milwaukee, WI, USA
Subscribe to:
Posts (Atom)